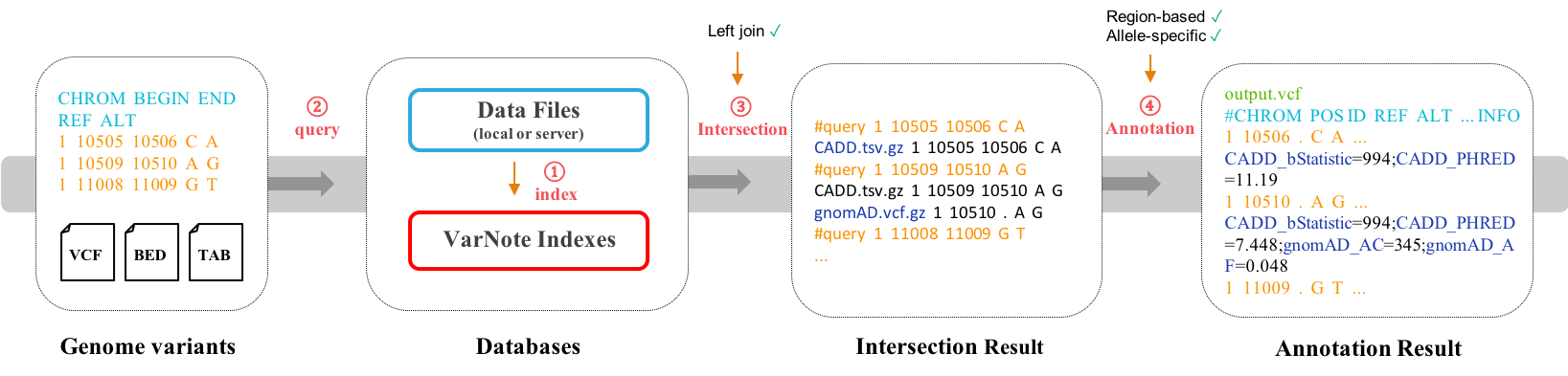
VarNote is a tool to rapidly annotate genome-scale variants from large and complex functional annotation resources.
VarNote is equipped by a
Genome-scale Variant Prioritization Applications
The advanced features of VarNote enable it can be extended to efficiently annotate and prioritize whole-genome non-coding variants in different genetic studies, such as GWAS downstream analysis and whole-genome sequencing of patients with inherited diseases. We leveraged VarNote programming APIs and large-scale variant annotations to provide three commonly-used applications, including
- VarNote-REG: prioritization of
likely causal regulatory variants from GWAS results; - VarNote-PAT: prioritization of
rare pathogenic regulatory variants from WES/WGS of patients with inherited diseases; - VarNote-CAN: prioritization of
cancer driver regulatory mutations from tumor sequencing results.
By integrating 127 Roadmap tissue/cell type-specific epigenomic profiles and 5 tissue/cell type-specific prediction tools (including cepip, GenoSkylinePlus, FUN-LDA, FitCons2, GenoNet), VarNote-REG can efficiently prioritize causal regulatory variants in the LD of each GWAS signal and provide combined scores.
By introducing VarNote filtrations on allele frequency, genetic inheritance model and genomic features, VarNote-PAT can significantly reduce the analytical time for WGS/WES data. Using combination scores, VarNote-PAT also showed improved performance in prioritizing true pathogenic regulatory variants of inherited diseases.
By annotating somatic recurrence of cancer non-coding regulatory mutations and incorporating our regBase-CAN cancer driver regulatory mutation prediction model, VarNote-CAN can prioritize likely cancer driver regulatory mutation given personal cancer genome profile.
Please cite VarNote as follows:
Huang D, Yi X, Zhou Y, Yao H, Xu H, Wang J, Zhang S, Nong W, Wang P, Shi L, Xuan C, Li M, Wang J, Li W, Kwan HS, Sham PC, Wang K, Li MJ*. Ultrafast and scalable variant annotation and prioritization with big functional genomics data. Genome Res. 2020 Dec;30(12):1789-1801.